Zhao Jincun’s Research Team and BGI Discovered Intra-host Variation and Evolutionary Dynamics of SARS-CoV-2 Populations in COVID-19 Patients
2021-02-231154Zhao Jincun’s Research Team of the State Key Laboratory of Respiratory Disease and BGI discovered intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients at different time points and in different anatomic sites, providing important theoretical guidance for the evolution of COVID-19 and the emergence of its mutations. Relevant paper Intra-host Variation and Evolutionary Dynamics of SARS-CoV-2 Populations in COVID-19 Patients was published online in Genome Medicine on February 23, 2021.

Figure 1 “Intra-host Variation and Evolutionary Dynamics of SARS-CoV-2 Populations in COVID-19 Patients”
Genome Medicine online
The study collected samples of COVID-19 patients at consecutive time points in multiple ways, including: nasopharyngeal swabs, sputum, feces, urine, and serosa, using transcriptome sequencing and probe hybrid capture to perform full-genome deep sequencing of COVID-19 at the same time, to compare and analyze the intra-host variation and evolutionary dynamics of viruses in different tissues of different individuals. The study found that the proportion of virus-related sequences of full-genome deep sequencing of clinical samples has a certain relation with PCR Ct values. Among them, virus-related sequences detected in respiratory tract (nose, pharynx, sputum) and intestinal samples (feces and anal swabs) are much higher than that in urine and serosa samples. 32 whole genome sequences were obtained through splicing. Compared with the reference sequence, 14 SNPs were found. A frameshift mutation was found in ORF8b, and the mutation of site diversity showed that the genetic variations of COVID-19 have obvious plasticity and adaptability. In addition, the study found that different branches of viruses co-existed in the intestinal samples of COVID-19 patients, which may be caused by coinfection with multiple viral subtypes, repeated mutations, or quasispecies changes.
Further, the research team conducted a mutational analysis of the same virus population (viral quasispecies) at different anatomic sites in the body, and found 40 intra-host single nucleotide variants (iSNV) in the individual, most of which (30/40) exist only in a single patient; while a small part (10/40) of which is shared among multiple patients. iSNV is an important tool for predicting the direction and bottleneck of the spread of the virus from person to person. Through comparative analysis, it is found that the distribution of iSNV has no significant relation with ORFs, codon position, and nonsense mutations, indicating that the current evolution of iSNV mainly relies on neutral mutation or incomplete purification. Most of iSNVs (30/40) detected are mainly rare iSNV (detected in only one patient), a small part of which (10/40) are common iSNVs (shared by multiple patients). Moreover, there is no obvious difference between rare iSNV and common iSNV in frequency of occurrence, indicating that the current type of COVID-19 has a severe genetic bottleneck. The comparison between SNP and iSNV also indicates that most of the iSNV currently detected cannot be effectively transmitted.
In addition, through comparison and analysis, the research team found that the diversity of COVID-19 in intestine is higher than that in respiratory tract. Respiratory tract samples and intestinal samples do not share the same iSNV, indicating that the there is less selection pressure in intestinal samples and variants are easier to occur. There are obvious genetic differences in two different anatomic sites. Longitudinal comparison of changes in the respiratory and intestinal populations at multiple time points found that iSNV in intestinal samples can be continuously detected at multiple time points, with obvious dynamic changes in frequency; while there are few continuous iSNV detected in respiratory samples, indicating that the mutations of COVID-19 in intestinal samples are more stable, the diversity of mutations is richer, and the mutation of intra-host single nucleotide variants in individuals is more sustainable.The above-mentioned intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients provide an important reference for the prevention and control of epidemic.
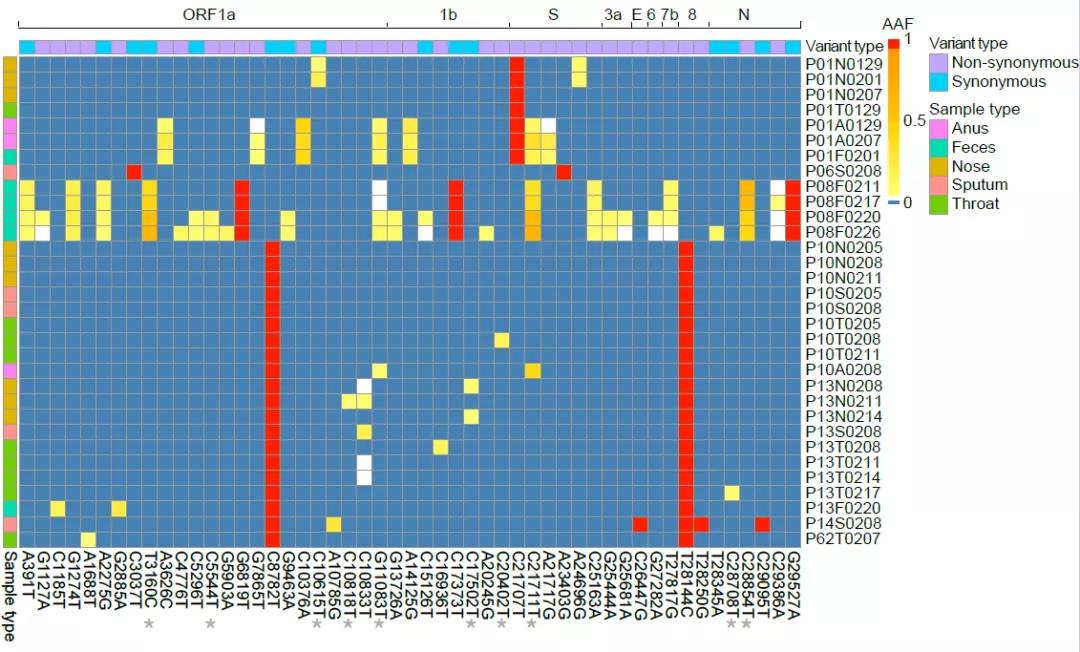
Figure 2 Distribution of iSNV in intraspecies virus population
The SARS-CoV-2 populations is the seventh coronavirus that infect humans discovered at the end of 2019. It is highly pathogenic and can spread quickly from person to person. As of mid-February 2021, it has infected more than 100 million people and killed more than 2.4 million people. Clarifying the evolutionary dynamics of COVID-19 is of great significance to the prevention and control of epidemic, pathogen detection, and vaccine development. The co-first authors of this paper are Wang Yanqun, Zhang Zhaoyong, Zhu Airu, and Huang Yongbo from the Laboratory, Wang Daxi and Sun Wanying from BGI, Chen Weijun from BGI PathoGenesis Pharmaceutical Technology Co., Ltd., and Zhang Lu from Guangzhou No.8 People’s Hospital. The corresponding authors are Professor Zhao Jincun and Professor Li Yimin from the Laboratory, and Professor Li Junhua from BGI. This project has received support from the National Science and Technology Major Project, the COVID-19 Special Project of the Ministry of Science and Technology, and the COVID-19 Special Project of Guangdong Province.
Original paper : https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00847-5
















