Analysis of New Coronavirus Mutations and Long Detoxification through Third-generation Nanopore Sequencing
2021-04-111341According to the World Health Organization, as of April 6, 2021, the COVID-19 pandemic has affected more than 215 countries and regions, causing 132,365,122 COVID-19 cases and 2,868,896 deaths worldwide. The new coronavirus is highly infectious. COVID-19 patients can be divided into asymptomatic, mild, moderate, severe and critical based on clinical manifestations and imaging examinations. Although previous studies have reported the genomic characteristics of the new coronavirus and the mechanism of the new coronavirus infection, it still needs to study the characteristics of the SARS-CoV-2 viral mutations carried by COVID-19 patients with different clinical manifestations, and their association and mechanism with the time of detoxification of the virus in the host.
With the in-depth research on COVID-19, based on the third-generation nanopore sequencing, the analysis results of the new coronavirus genome and its variants have been reported one after another. On July 9, 2020, Clinical Infectious Diseases (District 1, IF 8.313) published a case report entitled A Confirmed Case of SARS-CoV-2 Penumonia with Negative Routine Reverse Transcriptase – Polymerase Chain Reaction and Virus Variation in Guangzhou, China, which describes the advantages of the third-generation nanopore sequencing in positive rate of SARS-CoV-2 compared with RT-PCR, as well as its application in the clinical diagnosis of COVID-19 [1].

Third-generation nanopore sequencing can not only detect infections with multiple pathogens in one sample at the same time, but is also advantageous in positive rate compared with RT-PCR. It can also be used for the analysis of the new coronavirus genome, characterized by real-time data output and real-time analysis. In this study, researchers found SRAS-CoV-2 mutations in ORF10 based on nanopore sequencing, which was verified by first-generation sequencing, suggesting that this gene mutation may cause RT-PCR primer binding failure and negative COVID-19 virus test result.
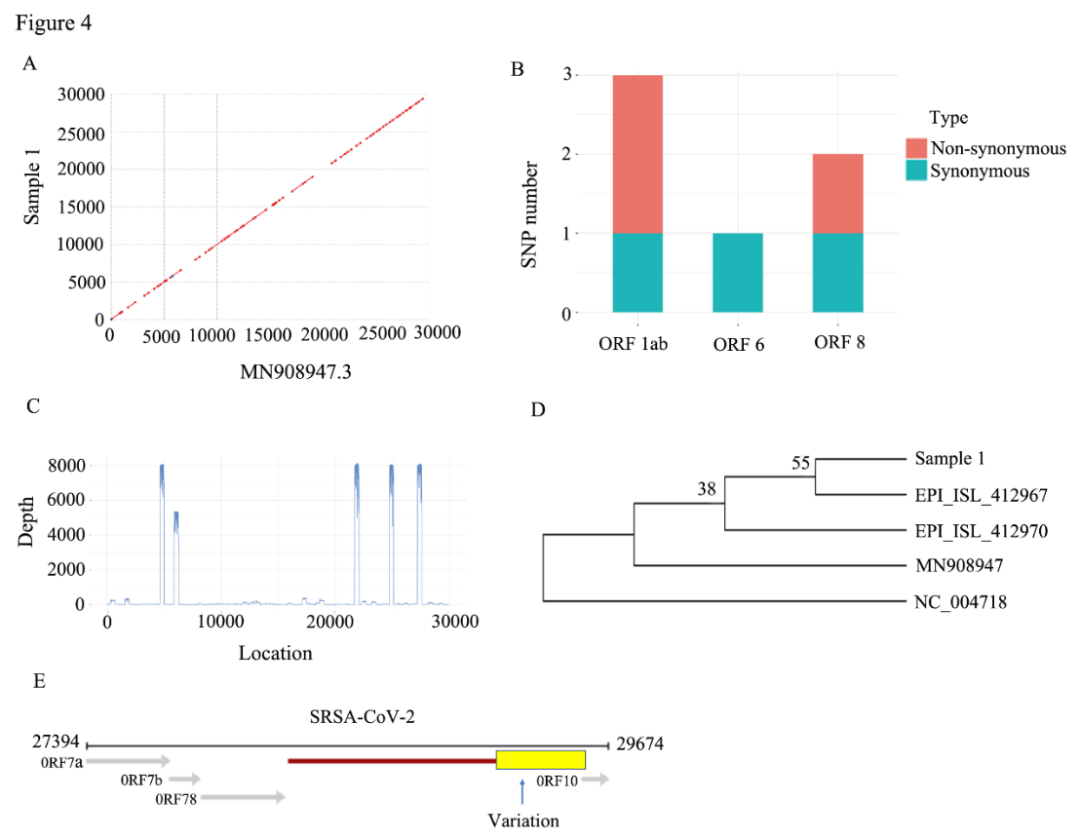
Figure 1: Analysis of mutations and evolutionary relationships of SARS-CoV-2 through nanopore sequencing
Based on previous studies, the research team used nanopore sequencing to analyze the gene mutations of SARS-CoV-2 and the host RNA expression in COVID-19 patients with long detoxification, in order to reveal the mechanism that leads to long detoxification and rejuvenation from the perspective of virome and transcriptome. The paper Longitudinal virological changes and underlying pathogenesis in hospitalized COVID-19 patients in Guangzhou, China has been officially accepted by Science China Life Sciences (IF 4.611), which is believed to be published soon [2]. The key findings of the study are summarized as follows:
1) The viral load in the lower respiratory tract of severe and critically ill COVID-19 patients is significantly higher; the clinical manifestations and SARS-CoV-2 positive detection time of some patients last more than 100 days. 2) Compared with patients with mild symptoms, the detoxification time of severe and critically ill patients is significantly longer, while the detoxification time of patients with retest positive results is longer. 3) Although the detoxification time of severe and critically ill patients is prolonged, the PCR test is still positive after the clinical manifestations disappear, but these patients will produce some specific neutralizing antibodies in their bodies, and no live SARS-CoV-2 has been detected in virus culture. 4) Long detoxification may be related not only to the gene mutations of A1,430G, C12,473T and G227A, but also to the overexpression of certain immune-related genes, such as IL1R1, IL1R2 and TNFRSF21 in the host.
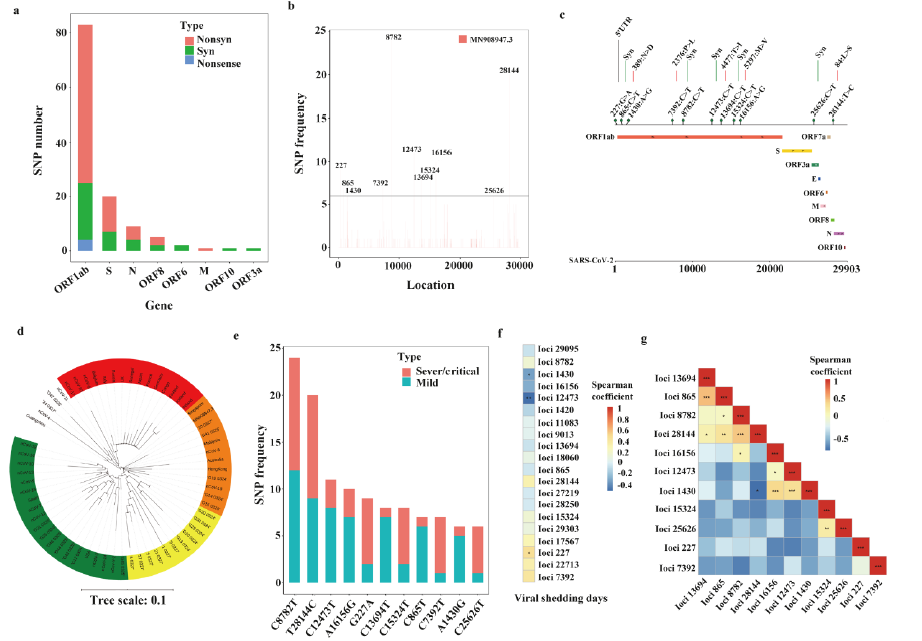
Figure 2: The SARS-CoV-2 gene mutations carried by patients with long detoxification
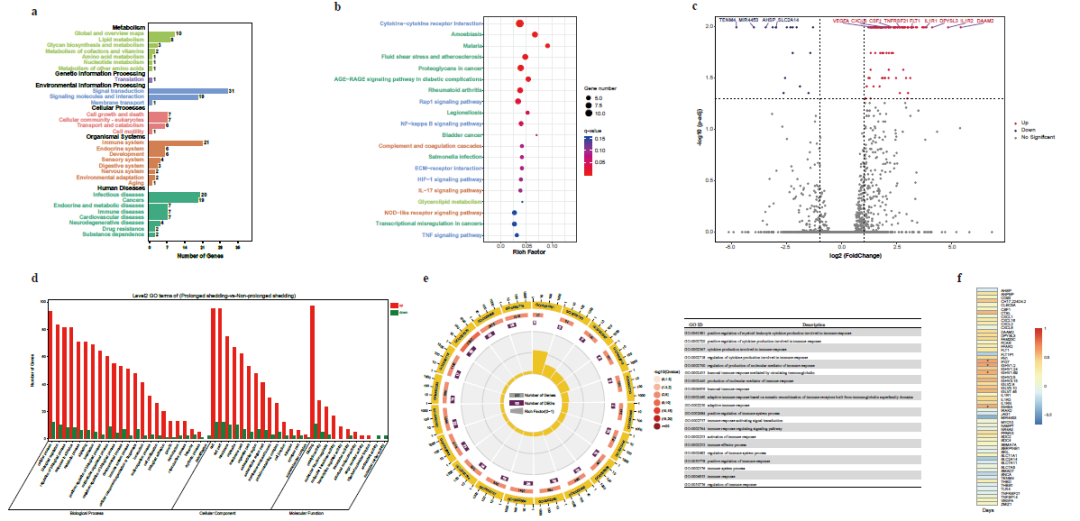
Figure 3: Gene expression in COVID-19 patients
The first author of the above two papers is Dr. Li Zhengtu from the Laboratory, and the corresponding author is Associate Professor PI Ye Feng from the Laboratory. The co-first units of the paper are the State Key Laboratory of Respiratory Disease, Guangzhou Institute of Respiratory Health, and the First Affiliated Hospital of Guangzhou Medical University. Meanwhile, a research team is formed to complete these studies by the Second People’s Hospital of Guangdong Province, City University of Hong Kong, and Beijing Yuansheng Kangtai Gene Technology Co., Ltd.
References
1 Zhengtu Li, Yinhu Li, Lingdan Chen, Shaoqiang Li, Le Yu, Airu Zhu, Feng Yang, Qian Jiang, Liyan Chen, Jincun Zhao, Wenju Lu, Nanshan Zhong, Feng Ye, A Confirmed Case of SARS-CoV-2 Pneumonia With Negative Routine Reverse Transcriptase–Polymerase Chain Reaction and Virus Variation in Guangzhou, China, Clinical Infectious Diseases, 2020
2 Zhengtu Li, Yinhu Li, Ruilin Sun, Shaoqiang Li, Lingdan Chen, Yangqing Zhan, Mingzhou Xie, Jiasheng Yang, Yanqun Wang, Airu Zhu, Guoping Gu, Le Yu, Shuaicheng Li, Tingting Liu, Zhaoming Chen, Wenhua Jian, Qian Jiang, Xiaofen Su, Weili Gu, Liyan Chen, Jing Cheng, Jincun Zhao, Wenju Lu, Jinping Zheng, Shiyue Li, Nanshan Zhong, Feng Ye. Longitudinal virological changes and underlying pathogenesis in hospitalized COVID-19 patients in Guangzhou, China. Science China Life Sciences, 2021 (Accept).
















